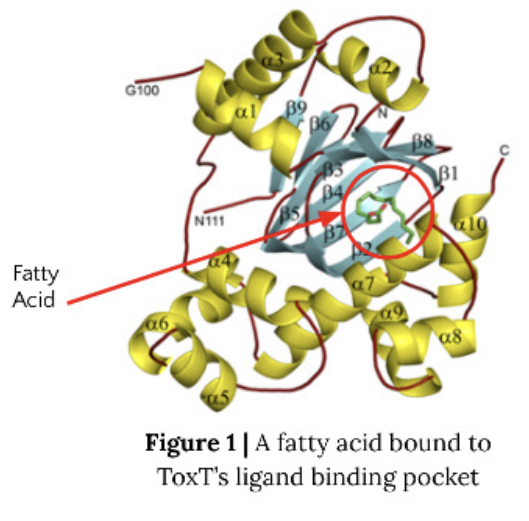
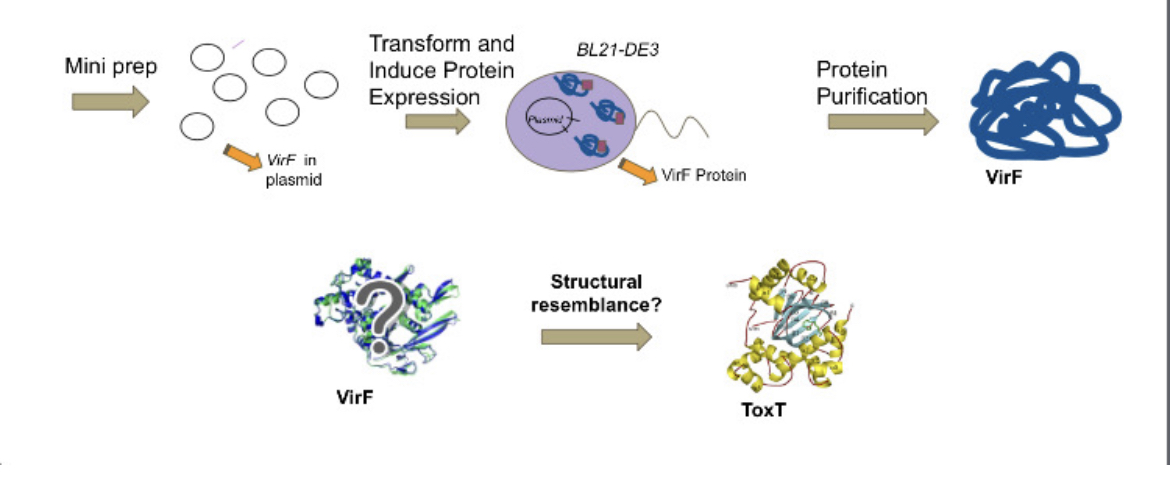
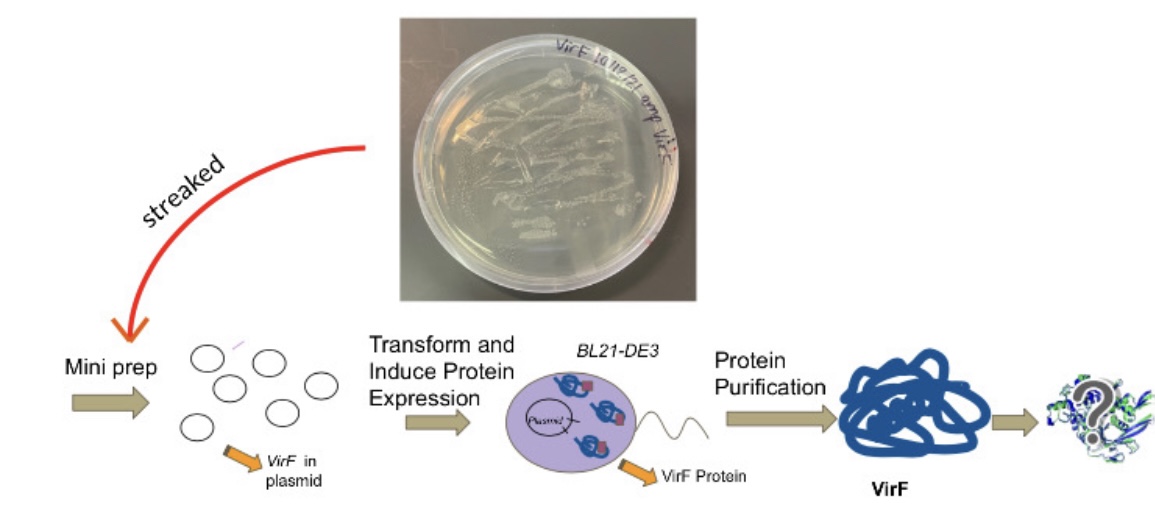
Shigella is a pathogen that is a leading cause of bacterial food-borne illness worldwide. The virulence cascade of shigellosis requires one protein of the AraC superfamily transcription factors, VirF. Other members of this superfamily, such as ToxT which is involved in the virulence cascade of Vibrio cholerae, show inhibition of DNA binding by a small fatty acid. We wish to determine whether or not VirF can be inhibited in a similar manner, and therefore we are working to express, purify, and ultimately solve the crystal structures of VirF. Solving the structures will help to reveal the mechanism of transcriptional regulation in S. dysenteriae and could lead to development of novel treatments for Shigella related illnesses. The Shigella bacterium causes shigellosis, specifically mammalian gastroenteritis. An estimated 80-165 million cases of gastroenteritis due to Shigella species occur globally each year. Symptoms vary from inflammation of the intestines causing diarrhea to nausea and vomiting. Shigella also kills over 600,000 people annually. Infection begins with the ingestion of contaminated food or water. Many of the genes required for intestinal penetration, invasion of host cells, and intestinal and diarrheal disease are carried out by the major regulatory protein VirF located on the PINV as part of the regulatory cascade. The member of the AraC protein family is a DNA-binding protein that activates toxicity factors icsA and virB genes. These genes are then able to activate other downstream effectors involving invasion of the cell wall and host infection. Thus, inhibiting VirF would inhibit the whole virulence cascade of shigellosis.