PETase 2022-2023 Project – Analyzing the Efficiency of Plastic-Degrading Enzymes
Maya Khan (Form V), Juliana Amorosi (Form IV), Sebastian Talarek (Form IV)
Project Summary + Project Goal
From overflowing landfills to islands of floating garbage in the oceans, trillions of pieces of abandoned plastic are contributing directly to a global pollution crisis. One of the most common types of plastic being polyethylene terephalate (PET). This synthetic material is a clear and lightweight plastic widely used for packaging. It’s known for its low price and convenient processability. However, the properties that make PET plastic so versatile also make it alarmingly resistant to biodegradation, giving it the ability to last for centuries. As discarded plastic is a direct contributor to environmental pollution, several technologies have been developed to treat plastic waste chemically, physically, and enzymatically. Unfortunately, many tested enzymes are limited by their required temperature of approximately 70℃ to carry out their activity. The bacterium Ideonella Sakaiensis, and its hydrolase enzyme PETase, was found to effectively use PET as a carbon source and can degrade the polymer at lower temperatures. Our project focuses on studying and optimizing the PETase enzyme by testing the effect of mutations on the enzyme’s efficiency and thermostability.
Current State
When we came out of Covid-19, we had to decide the future direction of the project. Our group had been given a complete list of mutations by another group of students at Greenfield Academy. We are focused on three (2 of which are mutants): Wildtype, PE3, and S238F. Our initial steps were transformation and autoinduction— done to induce protein expression within BL21 cells. This was followed by cell lysis: a protocol used to rupture the cell membrane and release intracellular contents. Then cell purification was performed to isolate the PETase enzyme. We ran several gels over the course of the year to check for the presence of PETase. Now, with our purified mutants, we plan to test for function.
Next Steps
In the upcoming year, we are going to develop a preliminary assay to test the enzymatic activity of PETase. This assay may include either a plate-clearing assay using E. coli cells or an in-vitro hydrolase assay. By establishing the baseline of enzymatic activity for our variants of PETase, we will be able to compare their activity to future PETase variants.
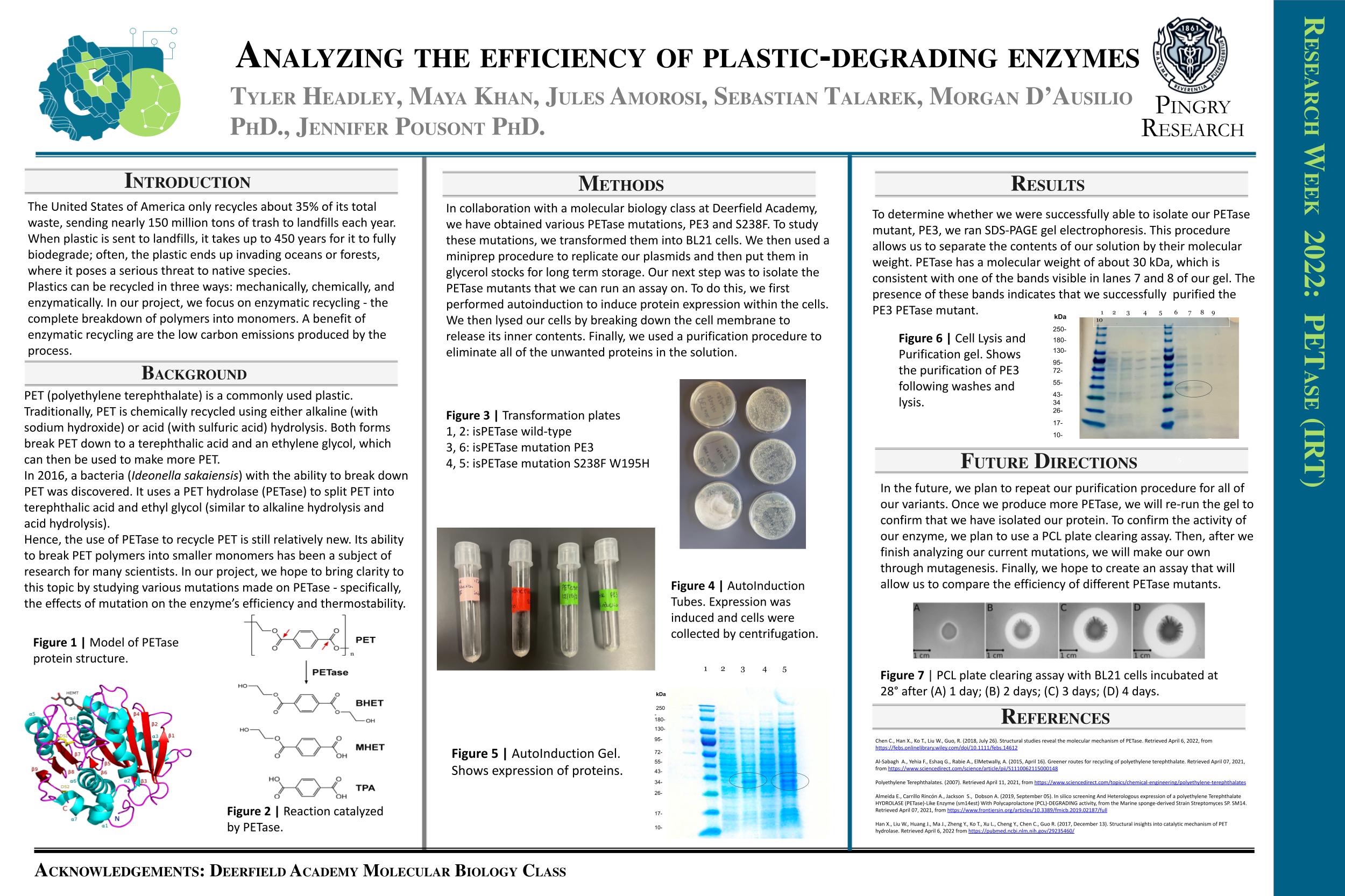
Research Week Poster
References
C., Han X., Ko T., Liu W., Guo, R. (2018, July 26). Structural studies reveal the molecular mechanism of PETase. Retrieved April 6, 2022, from https://febs.onlinelibrary.wiley.com/doi/10.1111/febs.14612 Al-Sabagh A., Yehia F., Eshaq
G., Rabie A., ElMetwally, A. (2015, April 16). Greener routes for recycling of polyethylene terephthalate. Retrieved April 07, 2021, from https://www.sciencedirect.com/science/article/pii/S1110062115000148 Polyethylene Terephthalates. (2007). Retrieved April 11, 2021, from
https://www.sciencedirect.com/topics/chemical-engineering/polyethylene-terephthalates
Almeida E., Carrillo Rincón A., Jackson S., Dobson A. (2019, September 05). In silico screening And Heterologous expression of a polyethylene Terephthalate HYDROLASE (PETase)-Like Enzyme (sm14est) With Polycaprolactone (PCL)-DEGRADING activity, from the Marine sponge-derived Strain Streptomyces SP. SM14. Retrieved April 07, 2021, from https://www.frontiersin.org/articles/10.3389/fmicb.2019.02187/full
Han X., Liu W., Huang J., Ma J., Zheng Y., Ko T., Xu L., Cheng Y., Chen C., Guo R. (2017, December 13). Structural insights into catalytic mechanism of PET hydrolase. Retrieved April 6, 2022 from https://pubmed.ncbi.nlm.nih.gov/29235460/
Meet the Team



Juliana Amorosi
Sebastian Talarek
Maya Khan

