Characterizing microbial community structure, dynamics, and succession in composters using metagenomics
Ryan Arrazcaeta, Ainsely Ellison, Dr. Sparrow
Project Summary
This project is dependent upon, and is being conducted in collaboration with, Ms. Olivia Tandon and the Composter Team.
Continuous drum-style composters contain diverse microbiomes which are crucial to nutrient cycling. Recent advancements in next-generation sequencing (NGS) technology have created unprecedented opportunities to characterize diverse microbial communities in complex environments. In combining time-course sampling with NGS technology, we can create a detailed “family portrait” of the composter microbiome. We also expect that sampling at multiple points within a composter will allow for comparisons between population profiles over time. These analyses will contribute a rich body of data to the comparatively new field of metagenomics. Finally, the sequencing of novel genomes may uncover microbes in biomass degradation that invite further research, such as in biochemical catalysis and biofuel production. A thorough characterization of the microbial communities in a composter would be both a project in general microbiology and an applied research investigation to improve our understanding of modern composters and operational efficiency.

Pingry composter with student, Ryan Arrazcaeta ’22, for scale.
Four Main Objectives:
This is a descriptive biology project that aims to identify the microbial communities present within a composter during startup and transition to a thermophilic regime. In doing so, this project aims to achieve four objectives:
1. Identify the presence and relative abundance of various microbial communities (bacteria, fungi, and archaea) present in a composter using next-generation sequencing as a composter starts up and its operating parameters change
2. Conduct physiochemical analyses to correlate changes in operating parameters to microbial community shifts
3. Identify and sequence novel species that are believed to play meaningful roles in composting/biomass decomposition after bioinformatic analysis
4. Describe the niches of different microbes in composters and their interactions with each other
Current State
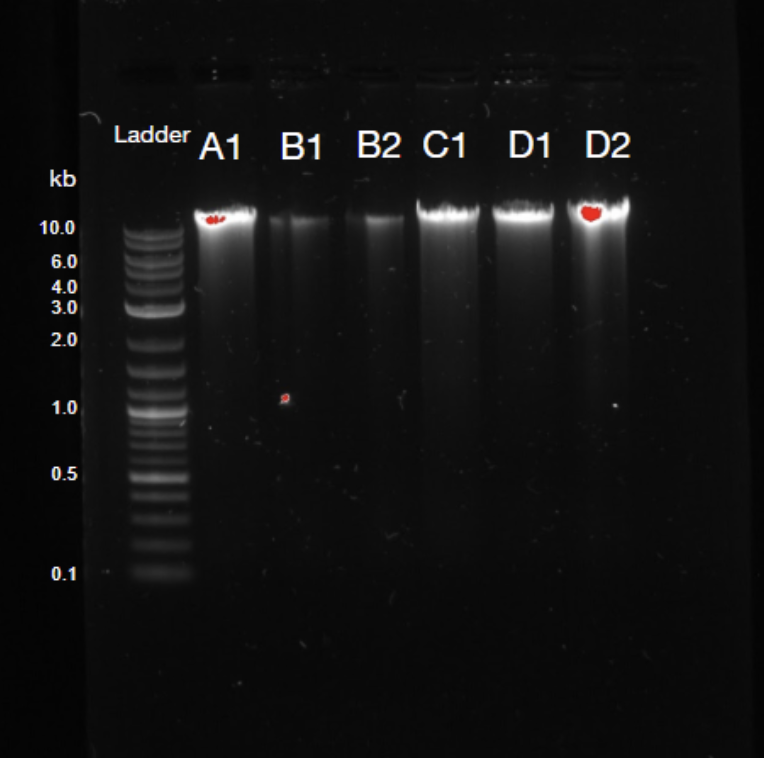
Figure 1. Agarose gel quality control. NGS requires long fragments for analysis, and streaks that are denser at the top of the gel are a positive sign; B1 and B2 are indicative of poor DNA extraction.
The Metagenomics Research Group was formed in 2019 and is an ongoing project. In recent months, we have studied the growing body of literature in the field of metagenomics. We have further begun to outline long-term project goals and protocols, in addition to conducting a series of preliminary trials. A pilot proof-of-concept trial confirmed our protocol’s reproducibility and produced reasonable results that can contribute to baseline data for future trials. Our initial analysis of composter DNA suggested the presence of previously uncharacterized species, and deeper sequencing of the DNA samples may allow for the identification and characterization of such species. Discoveries of this type have occurred following the bioinformatic analysis of microbial communities in naturally-occurring microbiomes. Due to the projected expenses of the project, we have worked towards securing funding from in-school and out-of-school grants.
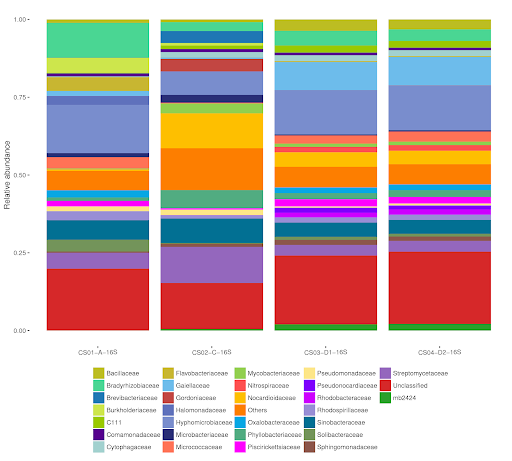
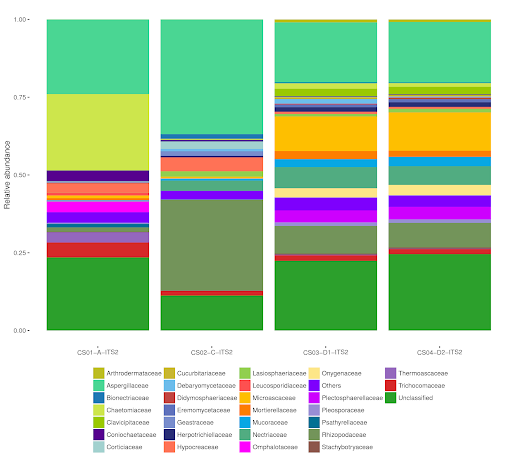
Figure 2. Example of NGS results for bacterial families.
Next Steps
Figure 3. Example of NGS results for fungal families
This project is still in its infancy. In the coming months, composter contents will be sampled from various points in the cylinder and at various times during the year. Temperature inside the composter will be monitored continuously using a submersible data logger. Samples will be analyzed for pH, moisture content, and carbon/nitrogen ratio. DNA will be isolated from dried and sifted samples and subjected to NGS of the generally accepted “barcode” genes 16S ribosomal RNA (for bacteria and archaea) and the ribosomal ITS sequence (for fungi). DNA will be isolated and assessed for quality at Pingry, while DNA sequencing and initial bioinformatic analysis will be outsourced to a contract lab.
Composter conditions will vary based on feedstock, position down the length of the composter, and time of year. Correlations will be sought between operating parameters and the microbial communities identified. The resulting data allow for longitudinal and cross-sectional comparisons through bioinformatic analyses. Special attention will be focused on (1) microbial communities that correlate with efficient composting and (2) the identification of novel species so as to create a cohesive “narrative” of the composter microbiome using sequencing results.
Meet the Team



